Pycorrelate examples¶
This notebook shows howto use pycorrelate as well as comparisons
with other implementations.
In [1]:
import numpy as np
import h5py
/home/docs/checkouts/readthedocs.org/user_builds/pycorrelate/conda/latest/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88
return f(*args, **kwds)
In [2]:
# Tweak here matplotlib style
import matplotlib.pyplot as plt
import matplotlib as mpl
mpl.rcParams['font.sans-serif'].insert(0, 'Arial')
mpl.rcParams['font.size'] = 14
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
In [3]:
import pycorrelate as pyc
print('pycorrelate version: ', pyc.__version__)
pycorrelate version: 0.3+24.gf9e109e
Load Data¶
We start by downloading some timestamps data:
In [4]:
url = 'http://files.figshare.com/2182601/0023uLRpitc_NTP_20dT_0.5GndCl.hdf5'
pyc.utils.download_file(url, save_dir='data')
URL: http://files.figshare.com/2182601/0023uLRpitc_NTP_20dT_0.5GndCl.hdf5
File: 0023uLRpitc_NTP_20dT_0.5GndCl.hdf5
File already on disk: data/0023uLRpitc_NTP_20dT_0.5GndCl.hdf5
Delete it to re-download.
In [5]:
fname = './data/' + url.split('/')[-1]
h5 = h5py.File(fname)
unit = 12.5e-9
In [6]:
num_ph = int(3e6)
detectors = h5['photon_data']['detectors'][:num_ph]
timestamps = h5['photon_data']['timestamps'][:num_ph]
t = timestamps[detectors == 0]
u = timestamps[detectors == 1]
In [7]:
t.shape, u.shape, t[0], u[0]
Out[7]:
((839592,), (1844370,), 146847, 188045)
In [8]:
t.max()*unit, u.max()*unit
Out[8]:
(599.9994419125, 599.9984722875)
Timestamps need to be monotonic, let’s test it:
In [9]:
assert (np.diff(t) > 0).all()
assert (np.diff(u) > 0).all()
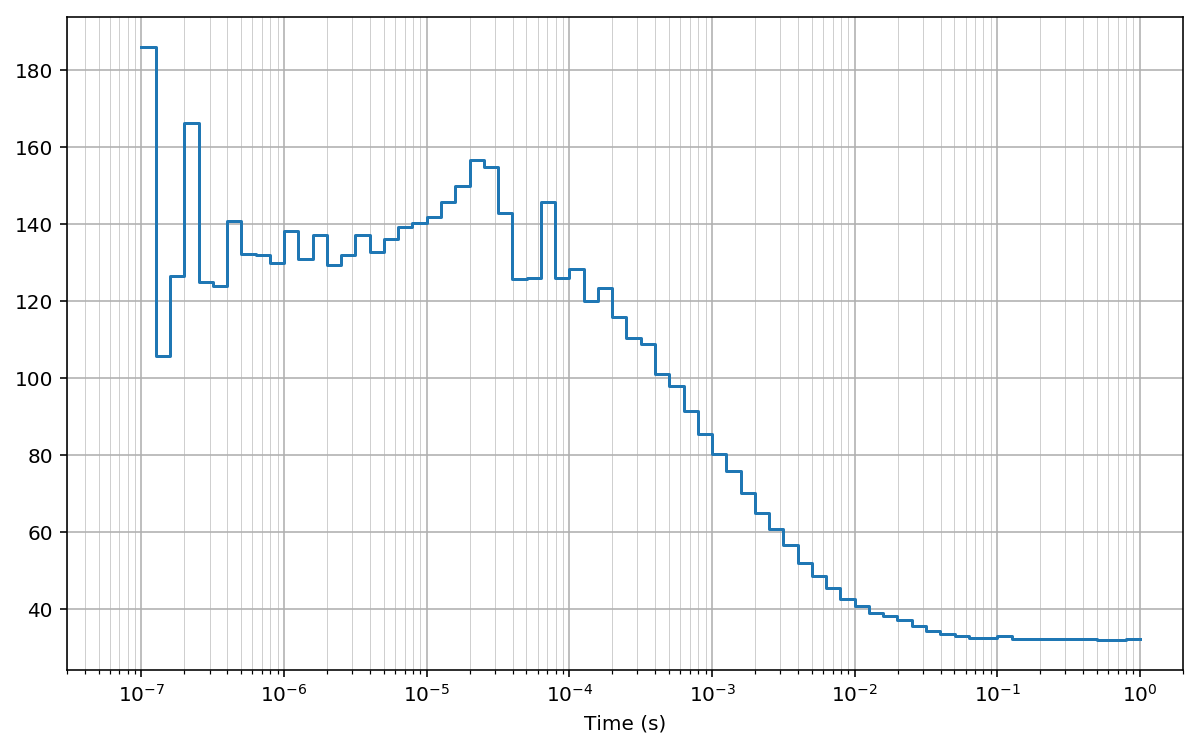
Log-scale bins (base 10)¶
Here we compute the cross-correlation on log10-spaced bins.
First we compute the array of lag bins using the function make_loglags:
In [10]:
# compute lags in sec. then convert to timestamp units
bins = pyc.make_loglags(-7, 0, 10, return_int=False) / unit
Then, we compute the cross-correlation using the function pcorrelate:
In [11]:
G = pyc.pcorrelate(t, u, bins)
/home/docs/checkouts/readthedocs.org/user_builds/pycorrelate/conda/latest/lib/python3.6/importlib/_bootstrap.py:219: RuntimeWarning: numpy.dtype size changed, may indicate binary incompatibility. Expected 96, got 88
return f(*args, **kwds)
In [12]:
fig, ax = plt.subplots(figsize=(10, 6))
plt.plot(bins*unit, np.hstack((G[:1], G)), drawstyle='steps-pre')
plt.xlabel('Time (s)')
#for x in bins[1:]: plt.axvline(x*unit, lw=0.2) # to mark bins
plt.grid(True); plt.grid(True, which='minor', lw=0.3)
plt.xscale('log')
plt.xlim(30e-9, 2)
Out[12]:
(3e-08, 2)
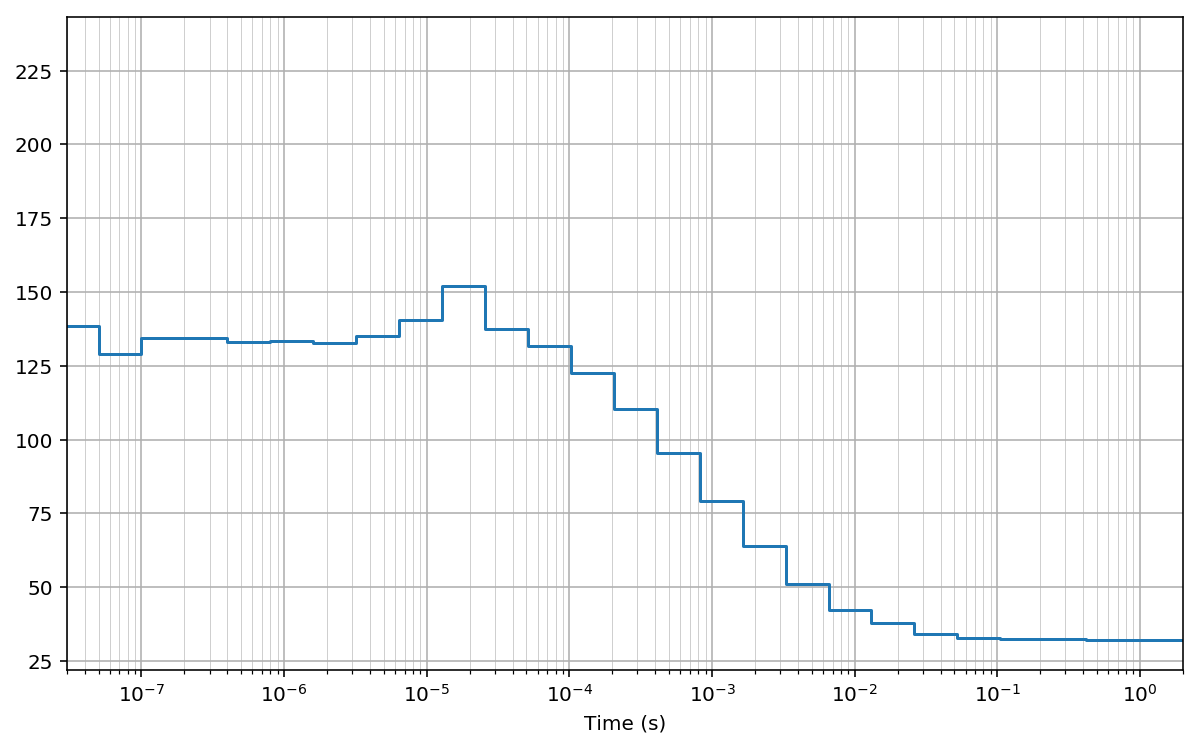
Log-scale bins (base 2)¶
Here we compute the same cross-correlation on log2-spaced bins.
First we compute the array of lag bins using the function make_loglags:
In [13]:
# compute lags directly in timestamp units
bins = pyc.make_loglags(0, 28, 1, base=2)
Then, we compute the cross-correlation using the function pcorrelate:
In [14]:
G = pyc.pcorrelate(t, u, bins)
In [15]:
fig, ax = plt.subplots(figsize=(10, 6))
plt.plot(bins*unit, np.hstack((G[:1], G)), drawstyle='steps-pre')
plt.xlabel('Time (s)')
#for x in bins[1:]: plt.axvline(x*unit, lw=0.2) # to mark bins
plt.grid(True); plt.grid(True, which='minor', lw=0.3)
plt.xscale('log')
plt.xlim(30e-9, 2)
Out[15]:
(3e-08, 2)
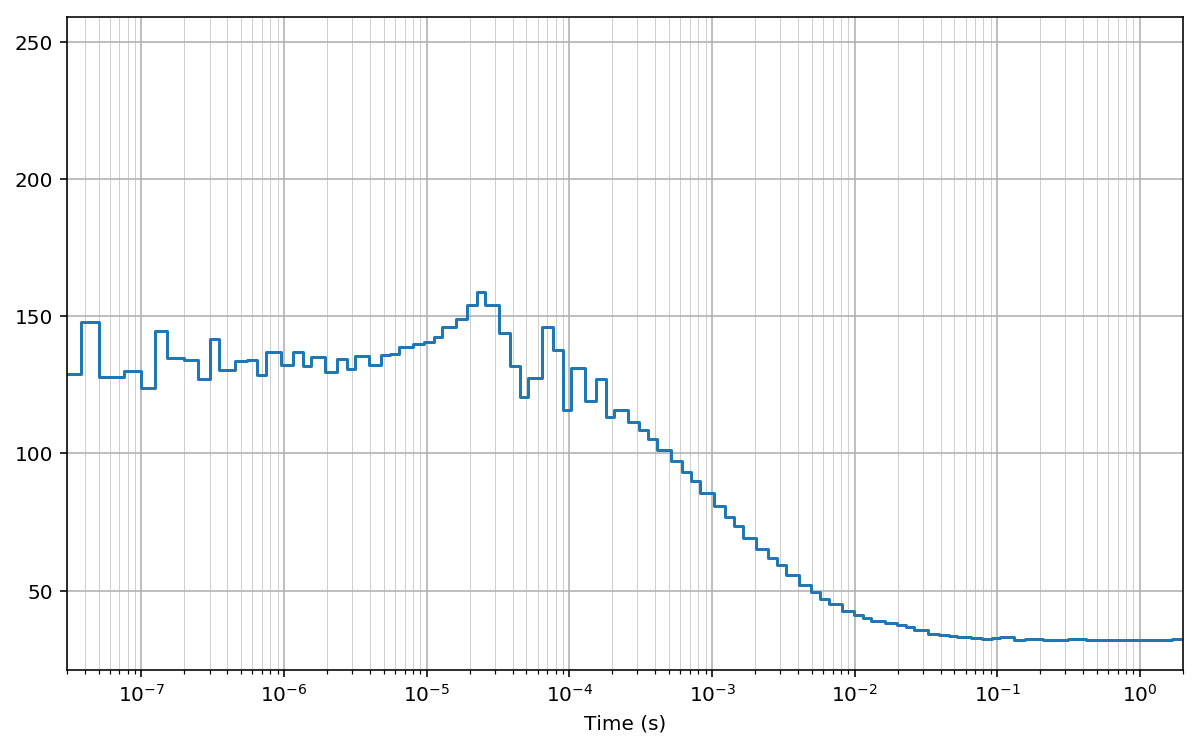
Multi-tau bins¶
Finally, we compute the cross-correlation on arbitrarly-spaced bins.
Similar to the multi-tau bins, here we use constant bin size for a
number of bins (n_group), then we double the bin size and we keep it
constant for another n_group and so on:
In [16]:
n_group = 4
bin_widths = []
for i in range(26):
bin_widths += [2**i]*n_group
np.array(bin_widths)
bins = np.hstack(([0], np.cumsum(bin_widths)))
Then, we compute the cross-correlation using the function pcorrelate:
In [17]:
G = pyc.pcorrelate(t, u, bins)
In [18]:
fig, ax = plt.subplots(figsize=(10, 6))
plt.plot(bins*unit, np.hstack((G[:1], G)), drawstyle='steps-pre')
plt.xlabel('Time (s)')
#for x in bins[1:]: plt.axvline(x*unit, lw=0.2) # to mark bins
plt.grid(True); plt.grid(True, which='minor', lw=0.3)
plt.xscale('log')
plt.xlim(30e-9, 2)
Out[18]:
(3e-08, 2)
Test: comparison with np.histogram¶
For testing alternative (slower) implementations we use smaller input arrays:
In [19]:
tt = t[:5000]
uu = u[:5000]
The algoritm implemented in pycorrelate.pcorrelate can be re-written
in a very simple way using numpy.histogram:
In [20]:
# compute lags in sec. then convert to timestamp units
bins = pyc.make_loglags(-7, 0, 10, return_int=False) / unit
In [21]:
Y = np.zeros(bins.size - 1, dtype=np.int64)
for ti in tt:
Yc, _ = np.histogram(uu - ti, bins=bins)
Y += Yc
G = Y / np.diff(bins)
In [22]:
assert (G == pyc.pcorrelate(tt, uu, bins)).all()
Test passed! Here we demonstrated that the logic of the algorithm is implemented as described in the paper (and in the few lines of code above).
Tests: comparison with np.correlate¶
The comparison with np.correlate is a little tricky. First we need
to bin our input to create timetraces that can be correlated by linear
convolution. For testing purposes, let’s use a small portion of the
timetraces:
In [23]:
binwidth = 50e-6
bins_tt = np.arange(0, tt.max()*unit, binwidth) / unit
bins_uu = np.arange(0, uu.max()*unit, binwidth) / unit
In [24]:
bins_tt.max()*unit, bins_tt.size
Out[24]:
(4.13725, 82746)
In [25]:
bins_uu.max()*unit, bins_uu.size
Out[25]:
(1.8020999999999998, 36043)
In [26]:
tx, _ = np.histogram(tt, bins=bins_tt)
ux, _ = np.histogram(uu, bins=bins_uu)
plt.figure(figsize=(10, 6))
plt.plot(bins_tt[1:]*unit, tx)
plt.plot(bins_uu[1:]*unit, ux)
plt.xlabel('Time (s)')
Out[26]:
Text(0.5,0,'Time (s)')
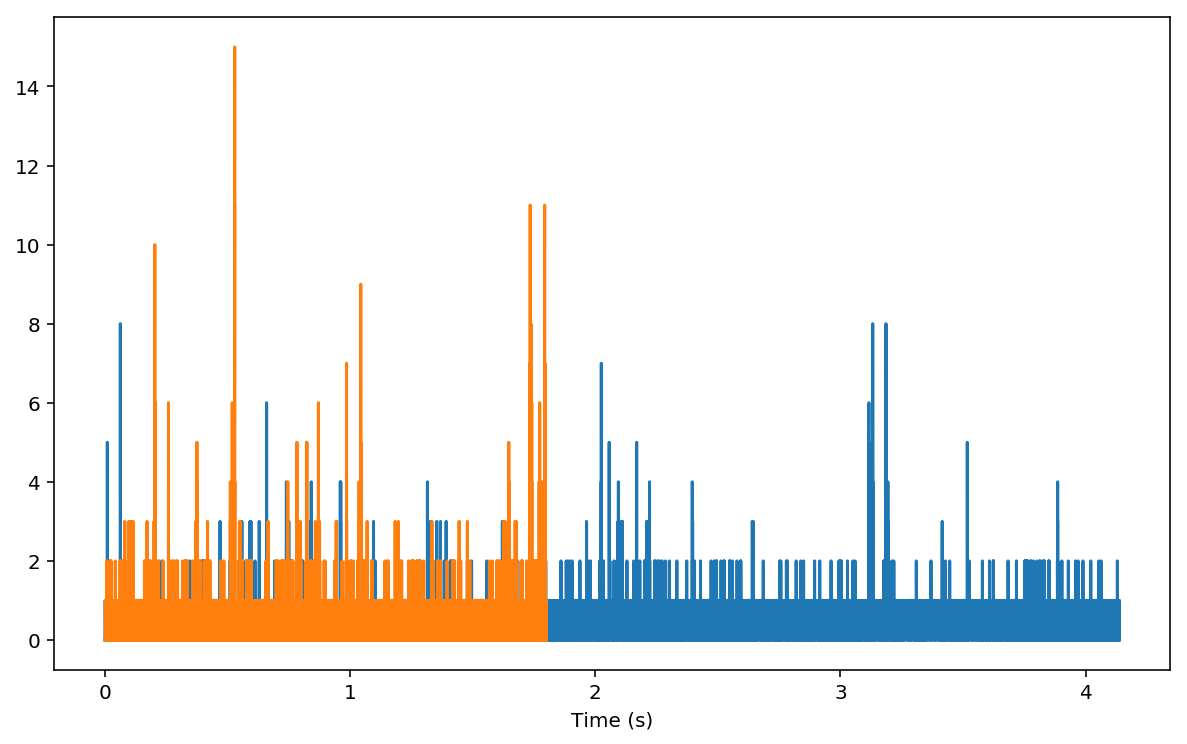
The plots above are the two curves we are going to feed to
np.correlate:
In [27]:
C = np.correlate(ux, tx, mode='full')
We need to trim the result to obtain a proper alignment with the 0-time lag:
In [28]:
Gn = C[tx.size-1:] # trim to positive time lags
Now, we can check that both numpy.correlate and
pycorrelate.ucorrelate give the same result:
In [29]:
Gu = pyc.ucorrelate(tx, ux)
assert (Gu == Gn).all()
Now, let’s compute the correlation also with pycorrelate.pcorrelate:
In [30]:
maxlag_sec = 3.9
lagbins = (np.arange(0, maxlag_sec, binwidth) / unit).astype('int64')
In [31]:
Gp = pyc.pcorrelate(tt, uu, lagbins) * int(binwidth / unit)
Let’s plot a comparison:
In [32]:
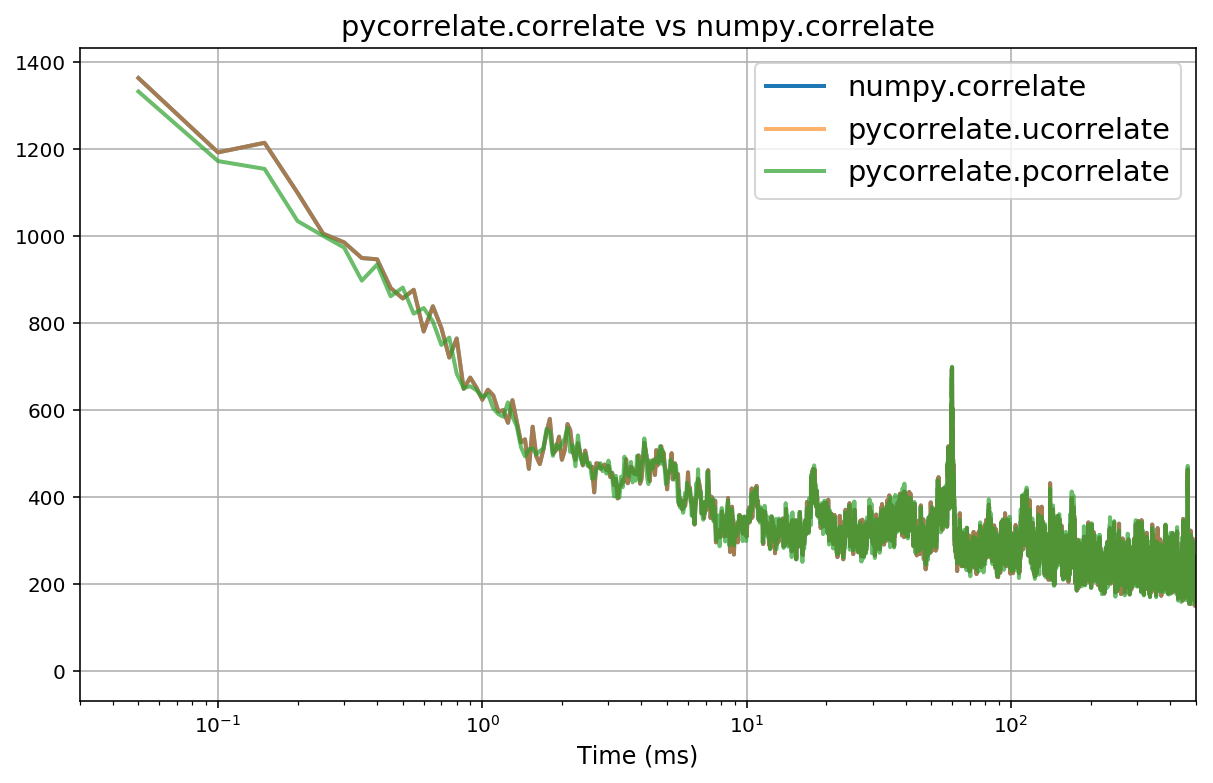
fig, ax = plt.subplots(figsize=(10, 6))
Gn_t = np.arange(1, Gn.size+1) * binwidth * 1e3
Gu_t = np.arange(1, Gu.size+1) * binwidth * 1e3
Gp_t = lagbins[1:] * unit * 1e3
plt.plot(Gn_t, Gn, alpha=1, lw=2, label='numpy.correlate')
plt.plot(Gu_t, Gu, alpha=0.6, lw=2, label='pycorrelate.ucorrelate')
plt.plot(Gp_t, Gp, alpha=0.7, lw=2, label='pycorrelate.pcorrelate')
plt.xlabel('Time (ms)', fontsize='large')
plt.grid(True)
plt.xlim(30e-3, 500)
plt.xscale('log')
plt.title('pycorrelate.correlate vs numpy.correlate', fontsize='x-large')
plt.legend(loc='best', fontsize='x-large');
Conclusion¶
numpy.correlateandpycorrelate.ucorrelategive identical results, with the latter being much faster. Note that the inputs are swapped between the two functions.pycorrelate.ucorrelateandpycorrelate.pcorrelateagree when using uniform time-lag bins.